IHCC Cohort Spotlight: Uganda Genome Resource (UGR)

Principal Investigator: Prof Segun Fatumo PhD, FHEA
The Uganda Genome Resource (UGR) is a well-characterized genomic resource that contains a variety of phenotypic communicable and non-communicable diseases and their risk factors derived from the Uganda General Population Cohort (GPC), a population-based open cohort founded in 1989. The UGR includes whole-genome sequence data on 2,000 Ugandan GPC individuals and genotyping data on 5,000 Ugandan GPC individuals representing ten ethnolinguistic groupings.
The GPC is a population-based study of approximately 22,000 individuals residing in 25 neighboring villages in the Kyamulibwa sub-county, Kalungu district in rural southwestern Uganda. The study was founded in 1989 by the Medical Research Council UK (MRC UK) in collaboration with the UVRI to study the epidemiology of HIV in a general population. The GPC was initially recruited and assessed through annual house-to-house census and survey rounds until 2012, when biannual surveys commenced. Since its establishment, more than 26 rounds of survey and 29 rounds of census have been undertaken. Before any survey procedures are carried out, written informed consent is obtained from participants on the use of their clinical records for research purposes and sample storage for future use. Data collected includes serological, demographic, and medical information from participants. Information regarding mortality, fertility, sexual behavior migration, and HIV infection perception are routinely collated.

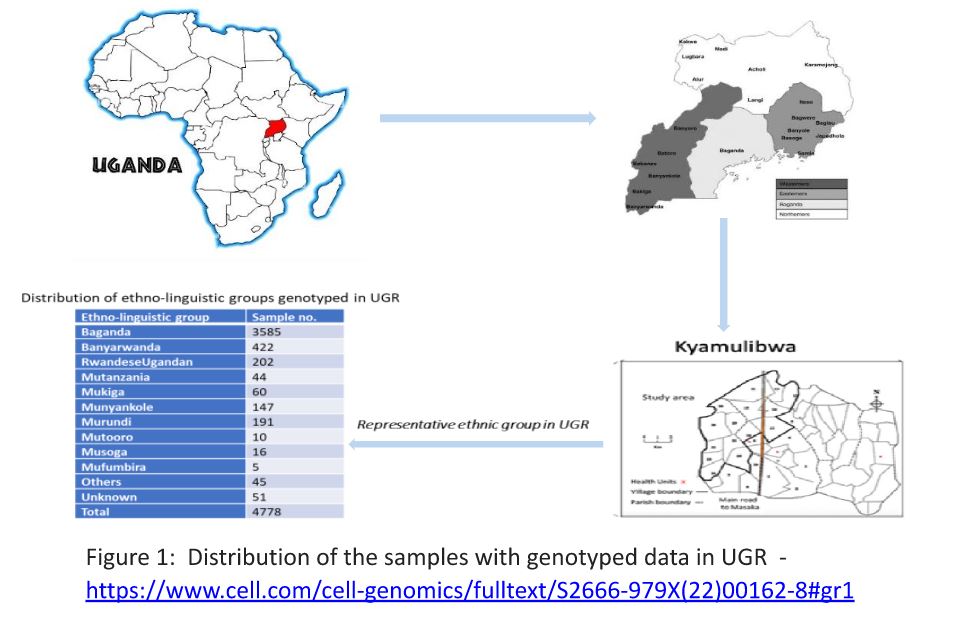
The GPC round 22 study of 2011 focused on the genetics and epidemiology of communicable disease and NCD, capturing different ethnic groups in Uganda for genomic studies (Figure 1). The 2.5M Illumina chip array was used to genotype nearly 5,000 Ugandans. The entire genomes of additional 2,000 Ugandans from 10 ethno-linguistic groups were sequenced using the Illumina HiSeq 2000 with 75 bp paired-end reads at low coverage, with an average coverage of 4× for each sample. All genotyped samples were imputed with a combined reference that was created by combining the UG2G sequence resource (n = 2,000, whole-genome sequence data from the African Genome Variation Project [n = 320]) and the 1000 Genomes phase 3 project (n = 2,504).
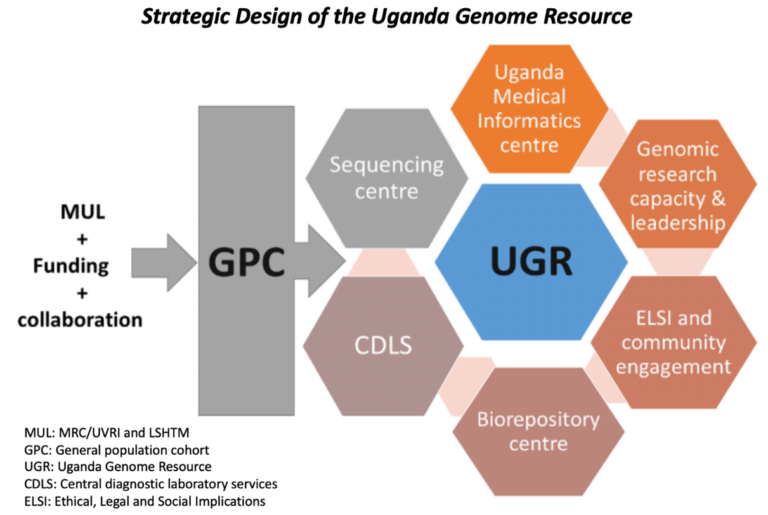
The cohort continues to function as a valuable platform for investigating the relationship between communicable illnesses and NCDs in a regular annual survey of the GPC. As shown in Figure 2, the UGR is supported by different platforms at the MRC/UVRI and LSHTM Uganda Research Unit (the Unit). The Unit has a reputation for leadership in genomics research capacity. The clinical diagnostic laboratory (CDLS) is an ISO 15189-certified laboratory that provides high-quality diagnostics testing support at the Unit. The Uganda Medical Informatics Centre (UMIC) is currently one of the largest health research-orientated computational resources in Sub-Saharan Africa with modern high-performance computing facilities to collect, store, and analyze data to advance genomic research. The Unit is also supported by a well-organized community engagement structure and activities, with strong support from the Community Advisory Board (CAB) including a biorepository for biospecimen storage and a DNA sequencing center.
Acknowledgments
We thank all participants who contributed to the Uganda Genome Resource. GPC was supported by the UK Medical Research Council ( MRC ) and the UK Department for International Development (DFID) under the MRC/DFID Concordat agreement, through core funding to the MRC/UVRI and LSHTM Uganda Research Unit. This work was supported by the Wellcome Trust grant number 220740/Z/20/Z to Segun Fatumo. Many thanks to the director of MRC/UVRI and LSHTM – Professor Pontiano Kaleebu, the deputy director – Professor Moffat Nyirenda and the current head of GPC – Dr. Joseph Mugisha for their support for UGR and GPC.
References
- Fatumo, Segun, Joseph Mugisha, Opeyemi Soremekun, Allan Kalungi, Richard Mayanja, Christopher Kintu, Ronald Makanga et al. “Uganda Genome Resource: A rich research database for genomic studies of communicable and non-communicable diseases in Africa.” Cell Genomics (2022). VOLUME 2, ISSUE 11, 100209, NOVEMBER 09, 2022 https://www.cell.com/cell-genomics/fulltext/S2666-979X(22)00162-8
- Gurdasani, Deepti, Tommy Carstensen, Segun Fatumo, Guanjie Chen, Chris S. Franklin, Javier Prado-Martinez, Heleen Bouman et al. “Uganda genome resource enables insights into population history and genomic discovery in Africa.” Cell 179, no. 4 (2019): 984-1002. https://doi.org/10.1016/j.cell.2019.10.004
- Asiki, Gershim, Georgina Murphy, Jessica Nakiyingi-Miiro, Janet Seeley, Rebecca N. Nsubuga, Alex Karabarinde, Laban Waswa et al. “The general population cohort in rural south-western Uganda: a platform for communicable and non-communicable disease studies.” International journal of epidemiology 42, no. 1 (2013): 129-141. https://doi.org/10.1093/ije/dys234